Tutorial
The web site provides service for user to upload protein structures to predict interaction sites for different types of ligands.
Prepare Input:
User can upload the file from the form. Up to 10 files can be submitted at the same time. The web server only recognizes protein structure in PDB file format (http://www.wwpdb.org/docs.html). Atoms start with HETATM will be ignored. For each file, the maximum size is 2MB.

User can also assign PDB ID and chain identifier if query structure is public available. The web will send query to the PDB website to retrieve the corresponding structure file.
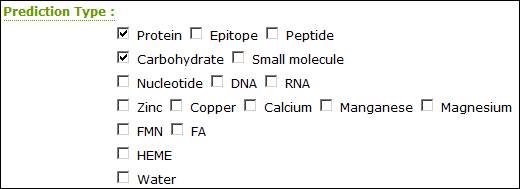
Selecting Predictors
Current available predictors are listed below. Use the check box to select multiple predictors.
Submit job and status query
Privacy
User can mark the job as private. Job with private mark will not be listed in job list. As the result, user can only retrieve the result through the link provide by the web.
E-mail Notification (Optional)
User can provide the E-mail address for notification when the job is finished. The E-mail address will not be used for other purpose.

Query job status
Once the job is submitted, the user will get a job id.
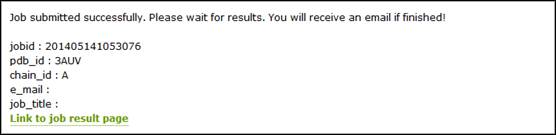
The link to the result will be refreshed every 30 seconds before the result is available.

User can also query the job ID or user defined job title through result page.
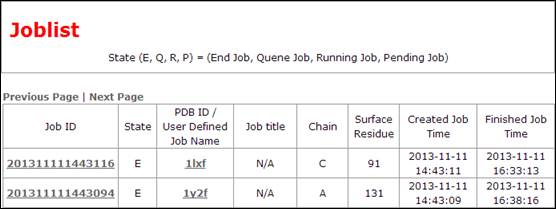
Joblist provides a list for all accessible results.
Result
Header
The header of each job shows the results of user selected predictors. For the example, the header includes protein-protein interaction and protein-small molecule (PL) and protein-peptide (PPep) predictors. User can switch the result between these predictions.

Display
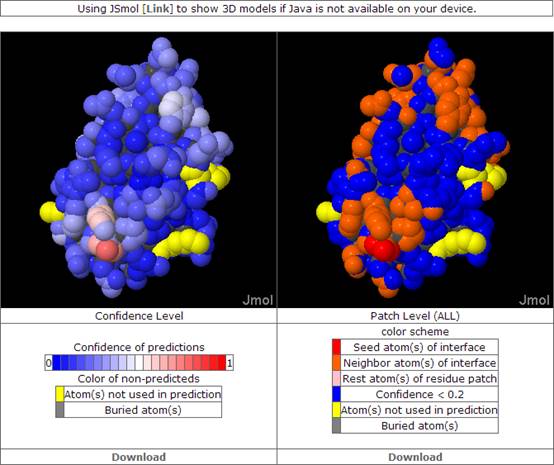
The predicted binding sites can be observed by using Jmol with Java enabled or JSmol if Java is not available on user’s device. The mouse actions on either applet will be synced to others if the checkbox of synchronize is checked. Two types of predictions are displayed, Prediction confidence level and predicting patches, are displayed in separate Jmol applets. Both confidence and patch results could be downloaded as PDB format file where B-factor field contains the corresponding confidence value or patch information. We also provide the details for each predicted patch and user can download the text file called PatchInfo. This PatchInfo file includes the atom serial, residue serial, residue type, predicted score and predicted type for all predicted patches. In addition to predicted result by NN with bagging, we also provide the predicted results by SVM (Support Vector Machine) and its output format is the same as NN output.
Prediction confidence level
Prediction confidence is based on the statistics of relationship between of machine learning output values and the percentage of these values in the binding sites calculated from the training set. Each surface atom of query protein will be assigned on numeric value ranged from 0 to 1 for representing the confidence of being binding site.
Predicting binding patch
Each binding patches were predicted as the following: For each of the atoms with confidence level greater than X, a circular patch centered at this atom with radius Z Å was mapped on the protein surface. Overlapped circular patches were merged into one patch. Each of the isolated patches was scored with the empirical function below:
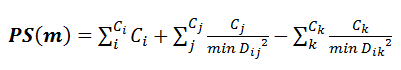
, where PS(m) is the patch score of patch m; Ci is the prediction confidence level of atom i in the patch m with prediction confidence level > X; Cj is the prediction confidence level ≤ X and > Y of atom j in patch m ; Ck is the prediction confidence level ≤ Y of atom k in patch m; minDij is the minimal distance of atom j to any atom i. minDik is the minimal distance of atom k to any atom i. By this PS score, we rank each possible rank and only top four patches are shown here. We also label each atom with color for indicating whether this atom belongs to predicted patch or not. Atoms with red color are predicted as binding atoms with confidence value >X and their structural neighbors with confidence level value which ≤ X and > Y are colored in orange..
Currently at most four patches ranked by score will be shown in the applet. User could also choose to display single patch by click the link.
Parameters used in predictors:
| Predictor Name | X | Y | Z Å |
| Protein-protein | 0.6 | 0.2 | 11 |
| Protein-carbohydrate | 0.5 | 0.1 | 5 |
| Protein-FMN | 0.7 | 0.2 | 9 |
| Protein-FA | 0.4 | 0.2 | 6 |
Sample page of result view.
Sequence based view
The prediction results can be mapped to sequence view. Confidence level of residue is mapped from its atom with largest confidence level. Predicted patches and residue based predictions are also colored separately.


